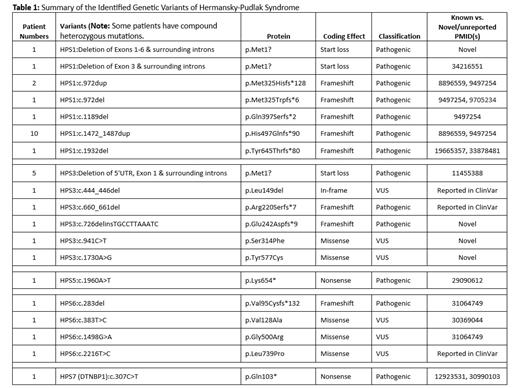
Background: Severe platelet (plt) dense granule (DG) deficiency is a hallmark of Hermansky-Pudlak syndrome (HPS), a rare, heterogeneous group of genetic disorders characterized by oculocutaneous albinism, bleeding diathesis, and lysosomal storage defects. A subset of HPS patients (pt) who has HPS1, HPS2, and HPS4 may also experience pulmonary fibrosis, granulomatous colitis, and/or immunodeficiency (syndromic HPS). Platelet transmission electron microscopy (PTEM) has been considered a valuable tool to screen such patients, and molecular testing aids in confirming te diagnosis, subclassifying the HPS and providing information on prognosis in pt. To elucidate the effectiveness of PTEM and next-generation sequencing (NGS) to identify and subclassify HPS pt, we conducted a comprehensive study on a cohort of pt with severe plt DG deficiency. Methods: After IRB approval, consecutive samples submitted to our reference laboratory for PTEM testing were analyzed. Whole mount (WM) PTEM tests were performed on all pt. Targeted NGS of the ISTH tier 1 genes (including 11 HPS-related genes) was performed on a cohort of patients who had < 0.3 mean DG/plt (200 plt counted) by PTEM. Data collected included demographics, PTEM findings, and genetic variants classified based on the 2015 ACMG criteria. Results: A total of 38 pt met our study criteria, the median age at PTEM testing was 39 years (range, 5 months to 76 years), and 22 (58%) were female. Ethnic backgrounds included Caucasian, Albanian, Puerto Rican, and other (unavailable). The mean DG/plt was 0 (0-0.2). A variant in an HPS-related gene was found in 26/38 patients (68%). The spectrum of variants in HPS-related genes is shown in Table 1, encompassing single-nucleotide substitutions, insertions, deletions, and frameshift mutations involving HPS1, HPS3, HPS5, HPS6, and HPS7 ( DTNBP1) genes. In addition to the previously reported pathogenic variants, several novel/previously unreported variants were discovered (Table 1). Interestingly, some patients presented with multiple variants, suggesting potential compound heterozygosity. No variants were found for the remaining 12 patients (32%). Conclusion: Our NGS-based genetic study of 38 patients with severe DG deficiency provides valuable insights into the complexity of this rare disorder. The high yield of finding variants in HPS-related genes verified that WM-PTEM is a useful screening tool for identifying HPS patients. The identification of known variants in HPS-associated genes confirms the importance of genetic testing not only in the diagnosis of HPS but also provides information on prognosis in syndromic HPS. Pathogenicity of novel variants remains to be elucidated. Moreover, the observed 32% of patients who had negative NGS results suggest the existence of as yet undiscovered genetic mechanisms for the severe platelet DG deficiency.
Disclosures
Pruthi:Genentech Inc.: Consultancy, Honoraria; Bayer Healthcare AG: Consultancy, Honoraria; Instrumentation Laboratories (Werfen): Consultancy, Honoraria; HEMA biologics: Consultancy, Honoraria; CSL Behring: Consultancy, Honoraria.